Signaling-to-transcription networks
Untangling the complex, biochemical networks cells use to drive specific transcriptional programs
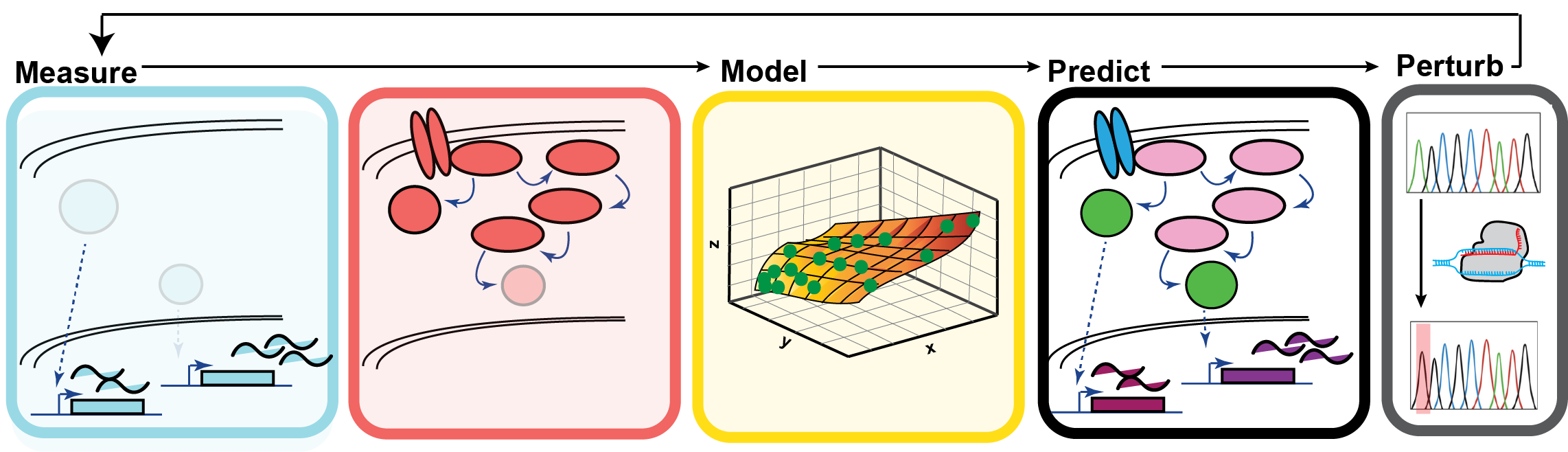
Understanding how cells interpret and respond to their environment is a fundamental goal of biology. Modern genomic and proteomic approaches have revealed the dynamics of thousands of post-translational modifications that may orchestrate the activation or repression of specific genetic programs. Our ability to map these intricate “signaling-to-transcription” networks and identify causal links between specific phosphorylation events and the expression of certain genes would not only better our comprehension of cellular decision-making, but will reveal a breadth of novel points of therapeutic intervention for us to modulate cell functions and behaviors.
We are developing and employing interdisciplinary strategies to build statistical models of signaling-to-transcription networks using global, temporally-resolved transcriptomic and phosphoproteomic data. We validate computationally predicted links between phosphosites and specific gene expression programs using conventional methods, as well as novel CRIPSR-mediated genomic engineering technologies to mutant individual PTM sites to assess their impact on gene expression at an unprecedented scale. The lab works mostly with embryonic stem cell models, and primary mouse and human T cells, as well as macrophages.